Table of Contents
Bleach Corrector
Author
Kota Miura
Bioimage Analysis & Research (BIAR)
Heidelberg
Video: Christophe Leterrier(@chrislet) See here for the twitter presenting this video
Citing the Plugin
- Paper: Miura K. Bleach correction ImageJ plugin for compensating the photobleaching of time-lapse sequences [version 1]. F1000Research 2020, 9:1494 https://doi.org/10.12688/f1000research.27171.1
- Code: Kota Miura et al. (2014). ImageJ Plugin CorrectBleach V2.0.2. Zenodo. 10.5281/zenodo.30769
History
- 10-09-15 First Version
- 12-04-16 Compiled with JRE 1.6.0_20
recent changes
- Bump to next development cycle (2023/02/22 15:28)
- Add license headers and project license (2023/02/22 15:27)
- POM: use HTTPS for schema location URL (2023/02/22 15:26)
- POM: fix forum.image.sc tag link (2023/02/22 15:26)
- Added the readme title (2023/02/06 10:10)
- Update README.md (2023/02/06 09:59)
- Merge pull request #8 from fiji/fix_exponential (2023/02/06 06:54)
- version 2.1.0 (2023/02/05 18:03)
- exponential method 2D, 3D corrected for 1 frame shift in estimation (2023/02/05 17:58)
- exponential method: added testing with main and verbose messages (2023/02/05 17:58)
Source
Released under the GNU General Public License v2.
Requires
ImageJ ver 1.34j or higher (ImageJ, upgrade page).
Installation
This plugin will be a part of Fiji, so you could find it under menu tree [Image > Adjust >] in near future (as of Apr. 18, 2012).
If you want to use it in ImageJ, download CorrectBleach_.jar (right click or ctrl-click to save) and place the jar file under plugin folder of ImageJ and restart ImageJ. You will find the plugin at [Plugins → emblTool → Bleach Corrector].
Description
This plugin contains three different methods for correcting the intensity decay due to photobleaching. They all work with either 2D or 3D time series. In case of 3D time series, image properties should be appropriately set. If you are not sure, check your image header by [Image → Properties].
- Simple Ratio Method:
- Plugin version of Jens Rietdorf's macro, extended further with capability for correcting 3D time series. This method is similar to the double normalization method in Phair et al. (2004), except that we do not normalize the curve.
- Phair, R. D., Gorski, S. A. and Misteli, T. (2004). Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol 375, 393-414.
- You need to estimate the base line intensity before using this method. Measure the mean intensity of the region outside the target signal and use that value.
- Exponential Fitting Method:
- Similar to the description in the manual of MBF-ImageJ. Additionally, this plugin also works with 3D time series.
- MBF-ImageJ suggests to use “Exponential” equation for fitting, whereas this plugin uses “Exponential with Offset”
- Figure below is an example of fitting exponential decay equation to the intensity changes over time. This is rather an ideal case example. If you see that the fit quality is not good enough, do not use this method. Beside the evaluation of the fitting quality by eyes, use R^2 (residual) as an indicator of the quality of fit.
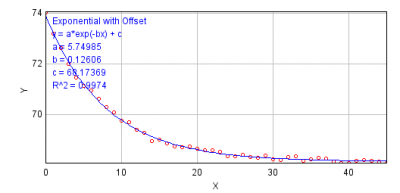
- Exponential Fitting Method (base subtraction):
- similar to above, but does two rounds of curve fitting.
- 1st round: estimate the baseline intensity and subtract the base line value
- 2nd round: do the curve fitting again and correct each frame accordingly.
- Histogram Matching Method:
- A brand-new method for bleach correction.
- This algorithm first samples the histogram of initial frame, and for the successive frames, histograms are matched to the first frame. This avoids the increase in noise in the latter part of the sequence which is a problem in the above two methods.
- This method does much better restoration of bleaching sequence for segmentation but not appropriate for intensity quantification.
- See the blog entry, a bit more detail on this issue and here is some more notes.
Headless Usage
This script demonstrates the headless usage.
Q & A
One of our users is making timelapse experiments to track a GFP marker in cell cultures. GFP signal is very dim and background is quite strong (so SNR very poor). Over the time, background intensity decreases while specific signal keeps more or less the same so it becomes gradually more visible. He really expects the GFP to increase over the time, and he would like to quantify this increase in GFP signal over time. To compensate background bleaching he is using your bleach_corrector plugin in FIJI. He obtains the best visualization of what he expects with the Histogram Matching Method. The thing is that, as you mention in your blog's entry (http://cmci.embl.de/downloads/bleach_corrector, http://cmci.embl.de/blogtng/2010-05-06/bleach_correction_2 ), with this method you cannot quantify intensities.
Why?
Can you recommend us an alternative method to be able to quantify changes in the GFP signal over time?
Other thing is at this moment it is difficult to know is wherther everything is bleached (so GFP signal kept constant reflects an increase) or wherther bleaching affects only the medium (so GFP is really constant and is not increasing, which is not what he expects…). We will make test to address this issue…
Xavier Sanjuan (ALMU, Parc de Recerca Biomèdica de Barcelona),
on behalf of Diego Barcena (Mark Isalan group, CRG, Barcelona)
The reason histogram matching cannot be used for the measurement, to explain in your case, is because the algorithm assumes that the histogram shape is always constant (which also means that the average intensity is constant over time).
On the other hand, you know that the signal should increase if its background is constant, which means that you must assume that histogram shape does change over time, contradicting with the assumption that histogram matching is based on.
One way that I can suggest to do the correction is as follows:
- Estimate the baseline intensity level.
- select a region (ROI) in the background, and do [Image > Stack > Plot z-axis profile…]. This will create a plot, expected to be decaying. Click “List” button which will then create a table with frame number and average intensity. Select All and Copy them all.
- Do [Analyze > Tools > Curve Fitting]. Curve fitting interface appears, so paste the copied value in the field (you probably need to delete the default values). Then fit the values using “Exponential with offset”
- You will then see a plot, fitted by a curve. the value “C” will be the estimate of baseline. Keep the value.
- Go back to your image stack, select a background region (ROI) again. This will be a region where the decay is measured.
- DO [Bleach correction], and select “Simple ratio”
- you will be asked for a background value. In put the value you got in 1.3. Click OK
- you will see the stack corrected by simple ratio method.