Analysis of FRAP Curves
Author: Kota
Miura (miura”a”embl.de), CMCI, EMBL Heidelberg, Tel. +49 6221 387 404
last update: April 29, 2010
For K_FRAPcalcV9jipf (IgorPro 6)
Features
-
IgorPro (Wavemetrics) Procedure Language (IgorPro ver.6 compatible)
-
Import FRAP measurement tab-delimited text files of
Zeiss data format (6 different data structures)
Leica data format (6 different data structures)
Olympus data formats
Excel data
CVS
- Three
different ways of normalizing FRAP curve
-
Curve fitting by single and double exponential formula and 2 diffusion formula.
-
Correction for the acquisition bleaching.
-
Output of half-max, diffusion coefficient, mobile/immobile fractions.
-
Weighting for the fitting.
-
Evaluation of goodness of fit by chi-square and gamma function.
-
Graphing of the fitted curve and estimation curve without acquisition
bleaching.
-
Batch analysis function for automated averaging of many curves.
Contents
1. Introduction
1.1. Qualitative
Interpretations
1.2. Quantitative
Interpretations
1.2.1. Chemical Interaction Models
1.2.1.2 Chemical Interactions with
immobile entity (not finished)
1.2.2.
Diffusion models (not finished)
1.2.3. Reaction-Diffusion Models
2. Analysis of FRAP
curves in Practice
2.1. Data Requirements
for Precise Measurements of Molecular Dynamics by FRAP
2.2. Terms
2.3. Normalization of Frap
Curve
2.3.1.
Normalization that involves acquisition
bleaching correction
2.3.2. Normalization that does NOT involve
acquisition bleaching correction
2.4. Determination of
acquisition bleaching parameters
2.5. Gap Ratio: Loss of
fluorescence due to the FRAP bleaching
2.6. Fitting Procedure
2.6.1
Single Exponential – Phair’s double
normalization
2.6.2 Single Exponential - Rainer’s Method
2.6.3 Single Exponential - Back Multiplication
Method
2.6.4. Phair’s Double Exponential Fitting
2.6.5. Soumpasis Diffusion Fitting
2.6.6. Ellenberg Diffusion Fitting
2.7. Evaluation of the Curve
Fitting: Goodness of Fit
3. Work Flow
3.1. Installing the FRAP Program
in the IGORPro
3.2. Importing Frap Data
3.2.1. Zeiss
3.2.2. Leica
3.3. Opening and Using Fit
Panel
3.4. Output
3.5. Batch Curve Fitting
Analysis
4. Appendix
5. References
1.1. Qualitative Interpretations
For a simple comparison of several different
experiment results, interpretation of FRAP (Fluorescence Recovery After
Photobleaching) curves can be qualitative. Just by looking at the graph the
speed of recovery to the plateau intensity can be examined. For example, Figure
1-1 shows several FRAP curves derived from cells under different treatments.
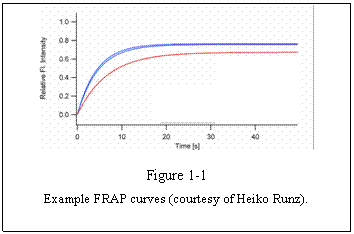 The recovery of blue curves is faster than the
red curve. We can then say that the mobility of the observed molecule is faster
in the blue curve condition than the red curve condition. The plateau level
seems to be also different in two conditions. Blue curve plateau is higher than
that of the red curve. We then know that the mobile fraction (see below) of the
molecule is larger in the blue conditions than in the red condition.
The recovery of blue curves is faster than the
red curve. We can then say that the mobility of the observed molecule is faster
in the blue curve condition than the red curve condition. The plateau level
seems to be also different in two conditions. Blue curve plateau is higher than
that of the red curve. We then know that the mobile fraction (see below) of the
molecule is larger in the blue conditions than in the red condition.
To add a bit more quantitative taste,
we can measure these two features (recovery speed and recovery fraction) numerically
from the FRAP curves. An example of actual FRAP curve after normalization is
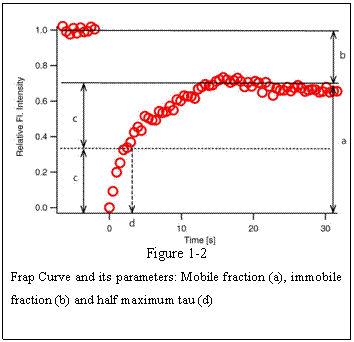
shown in Figure 1-2.
In many cases FRAP recovery curves do
not always reach the level of original fluorescence intensity. In Fig. 1-2 a, the
plateau level is lower than the pre-bleach fluorescence intensity because some
of the FRAP-bleached molecule are immobile within the FRAP ROI that they do not
contribute to the recovery and at the same time do not give away free binding
sites for incoming un-bleached proteins. For these reasons, fraction of proteins
that contribute to the recovery are called ‘mobile fraction’ and those do not are called ‘immobile fraction’. Each corresponds to a and b (=1-a) in figure 1-2,
respectively. Conventionally used index for the speed of recovery is the time
it takes for the curve to reach 50% of the plateau fluorescence intensity level
(c) and is called ‘half maximum’ or
‘half life’ and often abbreviated as
‘τ1/2’(d, tau half). Shorter half max tells us
that the recovery is faster.
A more objective way to extract half max
and mobile / immobile fraction is to fit the normalized FRAP curve I(t) by an exponential equation
![]() (1-1)
(1-1)
The fitted coefficients can be used
for calculating following parameters of the FRAP curve:
Mobile fraction = A (1-2)
Immobile fraction = 1-A (1-3)
Half Max: ![]() (1-4)
(1-4)
1.2. Quantitative Interpretations (Not Finished!)
For any kind of molecules within
biological system, two major factors affect their mobility. First is diffusion.
Diffusion of the molecule drives the change in the position of the molecule
from time to time. Rate of diffusion is determined by the diffusion constant of
the molecule, which is affected by the size of the molecule, viscosity and
temperature of the surrounding medium. Physical structures that hinder their
pure diffusion can also be considered as another factor and can be considered
as a parameter for the apparent viscosity of the medium (cf. Luby-Phelps). Second is the chemical interaction. The
environment of molecule is rarely a pure solvent in biological system. These surrounding
molecular species interact with the observed molecule. Binding and dissociation,
namely the binding constants of the molecule with others, affects the mobility
of the molecule. Therefore FRAP recovery curve is determined by two major
factors.
If the diffusion of the molecule is
fast and the interaction of molecule is slow, then the rate limiting factor for
the mobility is the interaction. In this case, we assume that the FRAP recovery
curve is dominated by the chemical interaction. If the interaction of the
molecule is faster than the diffusion, then the mobility of the molecule is
dominated by the diffusion.
Choice of equation for fitting the
recovery curve of FRAP experiments depends on what you assume for the mobility
of the protein that you are studying. There are three different models. (1)
Chemical Interaction Model (2) Diffusion Model (3) Reaction-Diffusion model.
1.2.1 Chemical Interaction Models
FRAP curves are fitted with exponential
equation in chemical interaction models,. In the simplest case, the equation is
with a single exponential term and called “single exponential equation” and was
already introduced above as the equation (1-1).
The exponential equation is an
analytical solution of a two-compartment model (Jacquez,
1972). We consider
molecule in one chamber A. The concentration of the molecule is [A] and the
molecule is moving to the neighboring chamber B though a narrow channel that
connects two chambers as shown in the illustration below (Figure 3.).
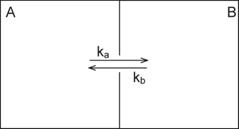
Figure 3.
Compartmental Model of a Simple Chemical Interaction
If the rate constant of transition
from A to B is ka and from B to A is kb, then the rate of the change of the
molecule concentration in the chamber B is
![]() (1-5)
(1-5)
Solving this equation using parameter variations and
assuming that [B]=0 when t=0, yields exponential equation (1-1) with
![]() (1-6)
(1-6)
![]() (1-7)
(1-7)
The interaction of observed molecule
is not necessarily limited to a single interaction. If the molecule we are
observing possesses two independent binding sites, then there are two
interactions.
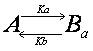
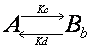
Although the interactions are
independent, we observe fluorescence recovery that is sum of these two
interactions and the analytical solution would be a double exponential equation
(see below).
Double exponential equation arises
also in other cases. For example, the interaction could be in two steps such
that the molecule interacts first weakly, and then upon this weak interaction
will there be a strong interaction.
![]()
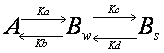
Ka,
Kb, Kc and Kd are rate constants. Although the state of interaction
is different, binding in weak and strong forms both contribute to the recovery
of the fluorescence. Thus the recovery curve is
![]() (1-8)
(1-8)
Since source of fluorescence outside
the FRAP ROI is enormous, we assume that [A]
is always constant.
![]() (1-9)
(1-9)
[Bw]
and [Bs] increases
with time. The rates of increase of the molecule with each binding state are
![]() (1-10)
(1-10)
![]() (1-11)
(1-11)
Solving these coupled differential
equations, we obtain a double exponential equation
![]() (1-12)
(1-12)
y0=A1+A2 (1-13)
1.2.1.2 Chemical Interactions with
immobile entity.
(Recovery depends only on Koff.
Refere to Sprague et al. 2004)
1.2.2. Diffusion models
There are two formulas proposed for
fitting the diffusion-limited FRAP recovery curves. Soumpasis modified the
initial formula proposed by Axelrod et al. (1974) as follows:
![]()
(Soumpasis, 1983)
This formula is purely theoretical.
Ellenberg et al. proposed an empirical formula as
below.
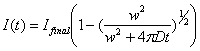
(Ellenberg et
al., 1997)
Details of the parameters used in
these equations will be discussed later. Note that the shape of the FRAP-ROI
affects the recover kinetics largely. Each fitting equation assumes certain
shapes for the FRAP-ROI. The area of the FRAP-ROI also matters so you must do
the experiment with a constant FRAP-ROI area.
1.2.3
Reaction-Diffusion Models.
Any molecular mobility has both
Reaction and diffusion components. Above two models are simplifies this reality
by assuming that one of these two components is negligible. Logically including
both components would be the ultimate direction of the modeling-based fitting (Sprague and
McNally, 2005; Sprague et al., 2004). This model is not
implemented yet in the program.
2. Analysis
of FRAP curves in Practice
2.1 Data Requirements for Precise Measurements of
Molecular Dynamics by FRAP
Following four different regions are
recommended to be measured for each experiment (see Figure 4).
(1) FRAP ROI (ROI=Region Of Interest). Region where you actually
bleached the fluorophore by strong laser irradiation. Shape and size of the
bleaching must be controlled. Shape of the FRAP ROI, circular, rectangular,
affects the model equation to be fitted with. Size of the ROI matters with the
diffusion-limited type of FRAP recovery (see below).
(2) Reference ROI Also
called ‘Cell Part’. ROI within the same cell (or it could be other cell) to
measure a decay in fluorescence due to the acquisition bleaching. The result
could be erroneous (see xx for more explanation).
(3) Base ROI Also called
‘background’. Set ROI outside the cell, where there should be no fluorescence,
to know the offset intensity.
(4) Whole Cell ROI or ‘All
cell’. The average fluorescence intensity of the whole cell you are observing.
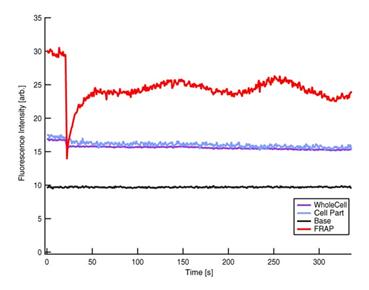
Figure 4
Fluorescence intensity kinetics at four different positions (courtesy of Mei
Rosa Ng).
Red: FRAP,
Blue: Part of cell, Purple: Whole Cell and Black: Base line
The minimal data set for measuring
the proper kinetics is FRAP ROI (1) and BASE ROI(4). It is possible to estimate
the recovery kinetics without (4), but if the baseline is high relative to the
signal and different from one another experiments, the interpretation of data
deviates largely from the reality. Complete set of data is all four different
ROI.
During experiment, we recommend to
take more than 20 frames before FRAP bleaching. This is because during the
initial acquisition phase fluorescence intensity bleaches largely and FRAP
bleaching is better be not overlaid to this systematic bleaching. In addition,
for the purpose of estimating “Gap Ratio” which indicates how much o total
fluorescence in the system you are observing is lost by FRAP bleaching,
pre-bleaching acquisition is better be longer. If you see flat fluorescence
intensity curve during pre-bleach, it should be fine. Furthermore, when the program
evaluates the goodness-of-fit, since standard deviation error of each time
point is in general cases not possible to estimate, the program uses
fluctuation of measured intensity during pre-bleach period to estimate
“measurement error” and is used for the calculation of gammaQ
value. For all these reasons, longer prebleach acquisition is strongly
recommended.
For numerical treatments of FRAP
recovery curves, several technical terms are used. See also Figure 5 and 6.
1. Intensity measurements from
different ROIs
FRAP ROI: Ifrap(t)
Reference
ROI: Iref(t)
Base
ROI: Ibase(t)
Whole Cell
ROI: Iwhole(t)
2. Intensity of FRAP ROI at bleaching
time point tbleach is
termed ’frap-bleach intensity’ or Ifrap-bleach.
Ifrap-bleach= Ifrap(tbleach)
3. Pre-bleach Intensities
Average fluorescence intensity before FRAP bleaching is
obtained by first subtracting the off-set fluorescence intensity (base ROI
intenisty) from other curves[1]
and averaging the value for pre-bleach period:
-
FRAP pre-bleach intensity: Ifrap-pre
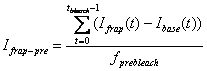
-
Reference pre-bleach intensity: Iref-pre
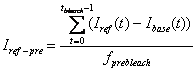
-
Whole Cell pre-bleach intensity: Iwhole-pre
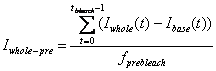
-
Whole Cell post-bleach intensity: Iwhole-post
Average intensity of the
Whole cell ROI after the bleaching. Use 10 time points after the
bleaching.
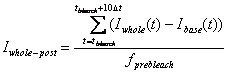
fprebleach is the frame (sampling) number
during pre-bleach period.
4. ‘
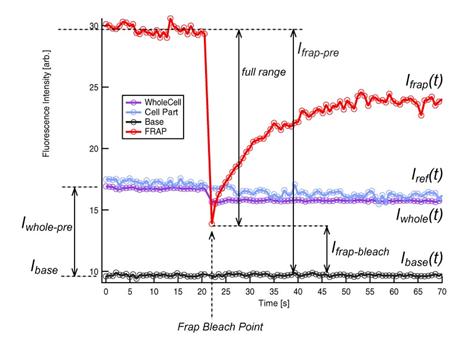
Figure 5 Terms used in FRAP recovery curve analysis
5. The ratio Iwhole-post
/ Iwhole-pre is called ‘gap
ratio’ (see Figure 6). This ratio is an indicator of the amount of
fluorescence that is lost form the cell by FRAP-bleaching. See section 2.6 ‘Gap Ratio:
Effects of FRAP bleaching on un-FRAPped region’ for details.
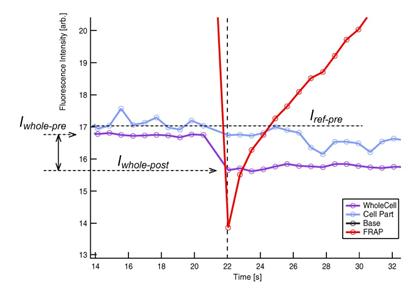
Figure 6 Terms used in FRAP recovery
curve analysis
2.3 Normalization of the Frap Curve
Any FRAP curve contains two
components of fluorescence intensity changes. The major component is the
recovery of fluorescence due to the flux of labeled proteins into the FRAP ROI.
The second component is the bleaching of fluorescence everywhere within the
microscope field where illumination light is irradiated to excite the fluorophore
for measuring their intensity –although very slow, acquisition bleaching causes
less and less molecules with fluorescence with time. This acquisition bleaching attenuates the fluorescence recovery, because
bleached molecules by this reason also move into the FRAP ROI and attenuate the
recovery of fluorescence. Thus, how to correct for acquisition bleaching is one
of the key point for the analysis of FRAP curve. In some cases, this correction
is included in the normalization procedure but in other cases correction is
independent from the normalization procedure. In this latter case, correction
will be done by including acquisition bleaching parameters to the FRAP equation
for fitting to the measured recovery curve. In the following, I will describe
different methods for doing normalizations.
2.3.1. Normalization that involves acquisition
bleaching correction This method is
known as ‘the double normalization’ (Phair et
al., 2004). In double normalization,
we take the measurement from whole cell ROI for correcting the acquisition
bleaching effects. Average pre-bleach whole cell intensity divided by the whole
cell intensity at each time points in the post-bleach period will be multiplied
to the FRAP curve at that time point. Before this operation, both Whole Cell
ROI and FRAP ROI data are subtracted by Base ROI intensity.
![]() (2-1)
(2-1)
In the ‘single normalization’ method
proposed by Phair et al., FRAP curve is simply subtracted by the offset
intensity outside of the cell (Base ROI), and then the base intensity is taken
as 0 and pre-bleach intensity Ifrap-pre of the FRAP
curve is taken as 1. If whole cell ROI or reference ROI data is not available,
then the single normalization method is employed but one should be aware that
this does not correct for the acquisition bleaching.
![]() (2-2)
(2-2)
In these methods, FRAP curves are
normalized to Ifrap-pre
that Ifrap-bleach >0.
For fitting equations that assumes Ifrap-bleach=Ifrap(tbleach)=0,
such as for diffusion-dominant models ‘Ellenberg’
and ‘Soumpasis’, (2-2) will be further
normalized to the full scale.
![]() (2-3)
(2-3)
2.3.2. Normalization that does NOT involve acquisition
bleaching correction The FRAP curve is
simply normalized by taking pre-bleach fluorescence intensity as 1 and bleach
intensity 0.
![]() (2-4)
(2-4)
In this method, Base ROI measurement
is not required. Acquisition bleaching will be corrected directly when the
curve is fitted, by using parameters for the acquisition bleaching
independently acquired through fitting the background decay curve. For more
detail, see ‘2.4 Determination of acquisition bleaching
parameters’. Following two methods uses this normalization method.
In ‘Rainer’s method’, the background
decay curve (either Whole Cell ROI or Reference ROI) will be normalized in the
same way as the FRAP ROI.
![]() (2-5)
(2-5)
This calculation may result negative
values for the normalized background decay curve but this does not affect the decay
parameter. In case of ‘Back Multiply’
method, normalization procedure takes two steps. First, normalize the background
curve by
![]() (2-6)
(2-6)
then normalize to the full range by
![]() (2-7)
(2-7)
2.4 Determination of acquisition bleaching
parameters
For obtaining parameters of
acquisition bleaching, we assume that the decay of fluorescence intensity
follows standard exponential decay.
![]() (2-8)
(2-8)
thus
![]() (2-9)
(2-9)
We fit the measured fluorescence
decay to the equation (2-9). As the t
approaches infinity, fluorescence level approaches to the base line level y0-decay since all fluorescence
will be bleached by time. In case when Whole Cell ROI data is available, then Iwhole(t) will be fitted. If
not, then Reference ROI data Iref(t)
will be used.
Figure 7
Different Patterns of exponential curves
One practical problem that you may encounter is fitting failure, which
will be reflected in the resulted parameters. There could be four combinations
for Adecay and τdecay. See Figure 7 for
the shape of each curve.
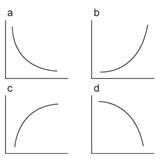
|
|
Adecay |
τdecay |
|
(a) |
+ |
+ |
|
(b) |
+ |
- |
|
(c) |
- |
+ |
|
(d) |
- |
- |
Among these combinations, the fitting
is successful only when both parameters are positive (a). (b) and (c) parameter
combinations results if the decay curve is an increase in the fluorescence
intensity by time, indicating that the quality of the measurement is bad. Double
negative parameters (d) results only when the curve has downward curvature,
which does not happen for the fluorescence decay by acquisition bleaching. For
these reasons, when the parameter combination is not double positive (a), the
program automatically sets the background flat and will be indicated in the
‘History’ window. One cause for such problems in the fitted curve could come from
the data close to the FRAP bleaching time point since they tend to be unstable.
You could avoid fitting those time points. Such an option is implemented in the
program (adjust “Back Start”).
2.5. Gap Ratio: Loss of fluorescence due to the
FRAP bleaching [2]
When relatively large part of the
sample is irradiated for FRAP bleaching, total fluorescence level within the
cell will decrease significantly (see Figure 6) by FRAP
bleaching. If you are also monitoring other part of the cell by Reference ROI,
although the decrease is more gradual, you may detect it as well, because
FRAP-bleached molecules moved to the Reference ROI and lowered its fluorescence
level (figure 6). The acquisition bleaching which you are observing then contains
two components. (a) Bleaching due to illumination light and (b) FRAP bleached
molecules moving into the ROI. Effect (b) is a complicated issue. The distance
between FRAP ROI and Reference ROI matters with kinetics e.g. closer the
distance between two regions, faster that you will detect the decrease.
Percentage of the fluorescence that you bleached for FRAP also matters. If the
FRAP-bleaching affects the decay of the reference ROI within the cell too much[3], you
need to obtain the acquisition bleaching kinetics from the whole cell. If the
effect of FRAP-bleaching on the reference point intensity dynamics is minimal
e.g. FRAP-bleached fluorescence is negligible against the total fluorescence of
the cell, then you can use the reference ROI for acquisition bleaching
correction.
We estimate the amount of
fluorescence loss due to the FRAP bleaching by obtaining the ‘Gap Ratio’. This value enables us to
calculate more precise mobile / immobile fraction of the molecule.
(1) When you have ‘Whole Cell ROI’
data[4]
If you have average fluorescence
intensity changes of the whole cell ROI, then the decrease fluorescence by FRAP
bleaching will be calculated by taking the ratio of Iwhole-pre and Iwhole-post.
![]() (2-10)
(2-10)
(2) When ‘AllCell’ data is NOT
available[5]
First, we estimate the decay kinetics
by fitting the equation (2-9) to post- FRAP bleaching period of Iref(t). Then we estimate the
average fluorescence level before the FRAP bleaching by extrapolating the fitted
decay curve[6]. This is an estimation
from the intensity changes after the loss of fluorescence by FRAP bleaching, so
we call this fluorescence level Iref-pre-est.
From the experiment, we know the measured fluorescence level before the FRAP
bleaching. We average these values and we call it Iref-pre-measured. Iback-pre-est
is smaller than Iback-pre-measured
because there is a loss in fluorescence due to the FRAP bleaching. Taking the
ratio between these two values, we obtain the Gap Ratio.
![]() (2-11)
(2-11)
We use this Gap Ratio for calculating
mobile and immobile fraction. The FRAP curve reaches plateau and we detect this
plateau level to know the mobile fraction. However, the plateau level never
becomes 1 because we bleached certain fraction of molecules by FRAP bleaching,
the fraction which is equal to the Gap Ratio. Hence, we calculate the actual
mobile fraction by
MobileFractionest =
MobileFractionmeasured / Gap Ratio (2-12)
2.6.1 Single Exponential – Phair’s double
normalization
After normalizing and correcting for the acquisition
bleaching, FRAP curve is fitted with standard exponential equation as explained
in the “chemical interaction dominant” model.
![]() (2-13)
(2-13)
At bleaching time point, I(tbleach) = y0+A
and the plateau of the curve is y0.
Mobile fraction will be calculated by
![]() (2-14)
(2-14)
and Half-Max calculation is same as
the equation (1-4).
2.6.2 Single Exponential - Rainer’s Method
The rate of fluorescence recovery is proportional to the available
binding sites within the bleached region. Simultaneously the recovery rate is
attenuated by fluorescence decay due to acquisition bleaching. This attenuation
is proportional to the fluorescence level at each time point. Taken together,
the rate of fluorescence recovery is
![]() (2-15)
(2-15)
Imax is a
fluorescence intensity corresponding to the maximum available binding site. Imax = Ifrap-pre
if all molecules are mobile. ![]() is a time constant
characteristic to fluorescence recovery, and
is a time constant
characteristic to fluorescence recovery, and ![]() is a time constant
that characterizes acquisition photobleaching. Solving the differential
equation (2-15) and taking that Ifrap(tbleach) = 0,
is a time constant
that characterizes acquisition photobleaching. Solving the differential
equation (2-15) and taking that Ifrap(tbleach) = 0,
![]() (2-16)
(2-16)
where,
![]() (2-17)
(2-17)
We obtain ![]() before these fittings by
fitting post-bleach period of Whole Cell ROI or Reference ROI data as explained
in details already (see 2.3) and use it as a
constant value during fitting. The estimation curve corrected for the
acquisition bleaching will then be
before these fittings by
fitting post-bleach period of Whole Cell ROI or Reference ROI data as explained
in details already (see 2.3) and use it as a
constant value during fitting. The estimation curve corrected for the
acquisition bleaching will then be
![]() (2-18)
(2-18)
For knowing the mobile – immobile
fraction, we consider the Gap Ratio as it was already explained (see 2.5 for more detail).
![]() (2-19)
(2-19)
Half-Max calculation is the same as
the equation (1-4) using![]() .
.
2.6.3 Single Exponential - Back Multiplication Method
The principle of correction for
acquisition bleaching is the same as Phair’s method, but instead of scaling
each time point of Ifrap(t)
by corresponding time point Iwhole(t)
or Iref(t), we use three
parameters of decay curve for directly fitting the un-corrected FRAP curve. The
advantage of this method over Phair’s method is that the amplification of error
due to multiplication of two data from each time point does not occur. Amplified
error will cause lowering of the Gamma-Q value for the evaluation of the
fitting (see 2.7). We fit the normalized,
uncorrected FRAP curve by following equation.
![]() (2-20)
(2-20)
![]() , y0, B
are obtained by fitting the decay curve, either Whole cell ROI or Reference
ROI, that is normalized by the procedure explained already (see 2.3). These three values are taken as constant
values during fitting. The estimation curve corrected for the acquisition
bleaching will then be
, y0, B
are obtained by fitting the decay curve, either Whole cell ROI or Reference
ROI, that is normalized by the procedure explained already (see 2.3). These three values are taken as constant
values during fitting. The estimation curve corrected for the acquisition
bleaching will then be
![]() (2-21)
(2-21)
Since decay curve parameters used in (2-20) is the
ones from already normalized decay curve, we do not need to consider the Gap
Ratio for knowing the mobile fraction.
Mobile = A (2-22)
Half-Max calculation is the same as
the equation (1-4) using![]() .
.
2.6.4. Phair’s Double Exponential Fitting
Similar to the single exponential
fitting, we fit the FRAP curve by
![]() (2-23)
(2-23)
At bleaching time point, I(tbleach) = y0+A1+A2
and the plateau of the curve is y0.
Mobile fraction will be calculated by
![]() (2-24)
(2-24)
2.6.5. Soumpasis Diffusion Fitting
We fit double-normalized FRAP curve
as explained already (see 2.3) to the
theoretical formula for diffusion FRAP curve
![]() (2-25)
(2-25)
where I0() is the modified Bessel function of the first kind
of order 0 and I1() is the
modified Bessel function of the first kind of first order to find only parameters
A and τ. A will be the mobile fraction and the diffusion coefficient
is calculated by
![]() (2-26)
(2-26)
where w is the radius (not diameter!) of the circular FRAP ROI and user
must know the value already. Formula 2-25 and 26 are taken directly from Sprague et al. 2004 and
definitions of parameters follow those of the paper[7]. In
the “FRAP” panel, you should input the radius in “FRAP width” field. I did not
use “FRAP radius” as the title of this field, since in case of Ellenberg
fitting, strip-bleaching is assumed and size of such fitting does not
correspond to “radius”. Half maximum of the recovery is estimated from the
estimation curve generated by the equation (2-25).
2.6.6. Ellenberg Diffusion Fitting
We fit double-normalized FRAP curve
as explained already (see 2.3) to the empirical
formula
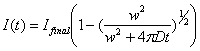 (2-27)
(2-27)
as proposed by Ellenberg et. al.
(1998). w is the width of the
strip-bleaching and need a user input. The fitting will look for a likely two
parameters Ifinal and D, the diffusion coefficient. Ifinal will be considered as
the mobile fraction. Half max can be calculated as
![]() (2-28)
(2-28)
2.7. Evaluation of the Curve Fitting: Goodness of
Fit
If data points are (xi,yi)
i=0,….N-1 and the model equation to fit is y(x)=y(x;a0….aM-1),
then the maximum likelihood estimate of the equation parameters is obtained by
minimizing the chi-square.
Measurement error σ can be obtained from the standard
deviation of pre-bleach FRAP intensities (ideal is standard deviation obtained
by repeating FRAP experiment, but in this program we treat pre-bleach standard
deviation as the constant measurement error). Using chi-square obtained by
equation (2-29), chi-square distribution for N-M degree of freedom can be
calculated using incomplete gamma function. Then this distribution gives the
probability Q that the chi-squared
should exceed a particular chi-square by chance[8].
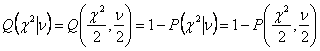 (2-30)
(2-30)
where υ =N-M is the number of
degree of freedom; N is the number of fitted points and M is the number of
parameters within equation to fit.
Following is an intuitive
explanation: Any measurement results deviate from ‘real values’ due to the
measurement error. Probability Q tells you if the chi-square calculated from
the fitting results are reasonably within the range of possible measurement
errors. Q > 0.1 can be considered a good fit, Q > 0.01 is a moderately
good fit, and Q < 0.01 recommends you either to think about different model
equation or to narrow down the range for the fitting[9] or
ultimately suggesting that something is wrong with the experiment. Q will be
printed in the history window as “gammaQ” value.
“…It is for this reason that reasonable experimenters
are often rather tolerant of low probabilities Q. It is not uncommon to deem
acceptable on equal terms any models with, say, Q > 0.001. This is not as
sloppy as it sounds: truly wrong models will often be rejected with vastly
smaller values of Q, 10-18, say. However, if day-in and day-out you
find yourself accepting model with Q ~ 10-3, you really should track
down the cause.”(numerical recipes in C)
In some cases, you might encounter
fitting results with Q=1, which sounds like a perfect fit. This is due to
over-estimation of measurement error, since larger standard deviation results
in smaller chi-squared value in (2-29) and hence would lead to better Q. If pre-bleaching intensity fluctuation is
large, then calculated Q for the fit becomes better because it tolerates larger
errors for the fitting. For this reason, if you encounter Q=1 but still need to
compare different fitting models for goodness-of-fit against a FRAP curve,
compare chi-squared values rather than Q values.
To see how gammaQ behaves, the graph
below plots gammaQ against different number of time points. For a fixed value
of Chi-sq, larger the number of time point indicates less error. For this
reason, gammaQ shows a sigmoid shape with 0 at lower N and 1 at higher N.
Increasing Chi-sq results in shifting the gammaQ value, meaning that the
evaluation results becomes more strict with same number of time points.
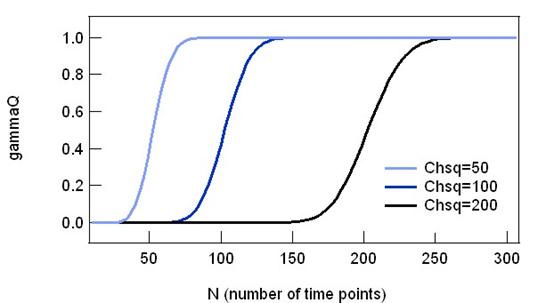
Fig. 2-7-1 gammaQ
behaviour
3.1. Installing the FRAP Program in the IGORPro
File à Open File à Procedure… à (select the file “K_FRAPcalcV9.ipf”) then click “Compile” at the
left-bottom corner of the opened window “K_FRAPcalcV9.ipf”.
You will see a new menu ‘Frap Calc’ after compiling the program (fig. 3-1).
Fig. 3-1 ‘FrapCalc’
menu
Zeiss and Leica each have their own data
output format. The program can import both formats, but the arrangement of data
is various. Four different types of fluorescence intensity sampling are possible
from FRAP experiments (see 2.1 and 2.2 for more detail) and how they are ordered depends on
the user. (Update: Olympus, Excel sheet, and Tab delimited data became
available to import , but will not add further explanation since the procedure
is similar to Zeiss or Leica)
(a) FRAP ROI time series
(b) Reference or Cell Part ROI time series
(c) Whole cell or All Cell ROI time series
(d) Base ROI time series
+
Data file also contains time point data. We call this “Time”.
The program has a list of typical
arrangements for each company’s data format[10].
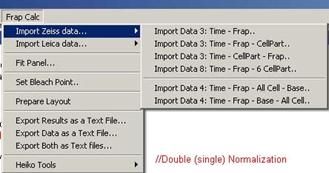
Fig. 3-2
Zeiss data importing
6 different types of data alignments
are possible now.
-. 2 columns: Time – Frap
-. 3 columns: Time – FRAP – Cell
Part
-. 3 columns: Time – Cell Part –
FRAP
-. 8 columns: Time – FRAP – 6 x Cell
Part
-. 4 columns: Time – FRAP – All Cell
– Base
-. 4 columns: Time – FRAP – Base –
All Cell
In Leica data format, data for each
ROI is separately written in different places within the file. Data first
starts with ROI1, with several columns containing frame number, time, channel1
and then channel 2 if there is. Then below ROI1, next ROI2 starts. For this
reason, one must select either single or double channel data structure.
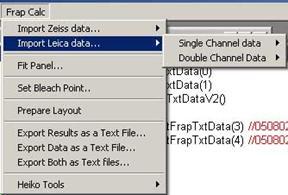
Fig.3-3 Leica
Data Importing
In case of single channel data, you
have two choice for the data structure.
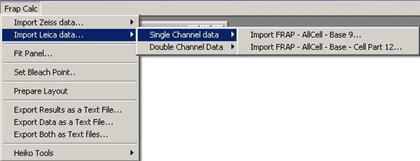
Fig. 3-4
-. single channel with three ROIs: FRAP – AllCell – Base
-. single channel with four ROIs: FRAP – AllCell – Base – Cell Part
In case of double channel data,
either channel 1 or channel 2 must be selected to tell the program which
channel is actually the frapped channel. If Channel 1 is the frapped channel then do
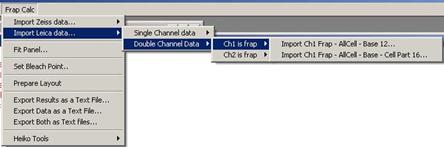
Fig. 3-5
If channel two is the frapped channel
then do
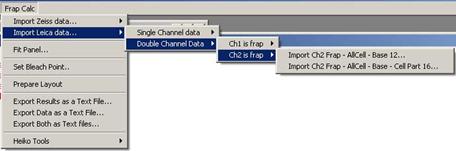
Fig. 3-6
In both cases , two types of ROI sets
are available.
- three ROIs: FRAP – AllCell – Base
- four ROIs: FRAP
– AllCell – Base – Cell Part
In all cases, when you select one of the importing command, pop
up window will appear (Fig. 3-7).
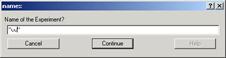
Fig.3-7
Input any name you like for the
experiment (but do not start the name with
a number & no space within the name!!). Click “Continue”. Another window
pops up to select the data file. Select, and then press OK. Almost immediately, a table and a graph like figure 3-5 will
appear. This is the original data. If you selected a data format that does not
matches with what you have selected in the menu, then importing is aborted and
a warning window appears, telling you how many columns the file contains.
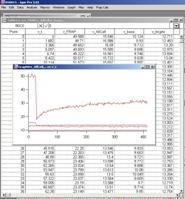
Fig. 3-8
Figure 3-8 shows a snapshot of screen
when you import Leica data file with full set of ROI.
3.3. Opening and Using Fit Panel
3.3.1. Open Fit Panel Frap calc à Fit Panel… This will create a panel
shown in the figure 3-6.
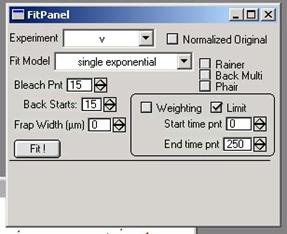
figure 3-6
3.3.2. Most important step before actual fitting is user-input of the
FRAP bleach point. Check through the table that was automatically opened when
you import the file and find out the point number (not time) of the bleaching.
Note that the numbering of cells start from 0, so 6th cell would be cell number
5, and if that is the time point of bleaching, you should set the bleaching
time point to 5. Then input the number at the “Bleach Pnt” field[11]. You
can toggle the number by clicking the up-down arrow at the right side of the
input window.
3.3.3. Using drop-down menu titled ‘experiment’ at the top of the panel,
select one of the experiment you imported and named.
3.3.4. Using drop-down menu for ‘Fit
Model’ at the second row, select one of the model that you are going to fit.
Four different models are possible.
-
Single exponential chemical reaction model
-
Double exponential chemical reaction model
- Ellenberg’s
diffusion formula
3.3.5. From three different normalization methods, choose one by
checking one of the check boxes beside the “Fit model” drop-down menu. Three
different normalization methods are possible:
-
Phair’s method (Double or Single
normalization)
Note: Check buttons will be enabled
or disabled automatically depending on the fit model you selected. For example,
single exponential model enables all three normalization methods but diffusion
models are only possible through Phair’s double normalization.
3.3.6. When you select Diffusion models, you must enter the “Frap
Width”, the size of the FRAP bleached area in µm. Otherwise resulting diffusion
coefficient will be wrong.
3.3.7. Press ‘Fit!’ button at the left-bottom corner of the panel. This
will create a new graph (if there is already a graph window for the
corresponding experiment, pressing the button will bring that window forward)
and do the fitting. The result is as shown in the figure 3-7 (Rainer’s – Single
Exponential), figure 3-8 (Back Multiply – Single Exponential), figure 3-9
(Phair – Single Exponential).

Fig. 3-7 Rainer’s
– Single Exponential Blue dots are the
decay curve (both Whole Cell and Reference data), Red dots are the FRAP data.
Light blue curve is the fitted FRAP curve. Yellow dotted curve shows the
estimation curve (see 2.x for details). At the top of the graph, residuals of
the fitting are plotted. Inadequate model will result a systematic deviations,
as you can see in this figure.
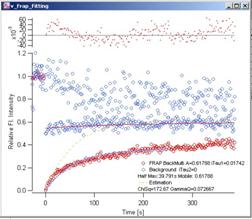
Fig. 3-8 Back
Multiply – Single Exponential Blue
dots are the decay curve (both Whole Cell and Reference data), Red dots are the
FRAP data. Red curve shows the fitting of whole cell data. Light blue curve is
the fitted FRAP curve. Yellow dotted curve shows the estimation curve (see 2.x
for details). At the top of the graph, fit-residuals
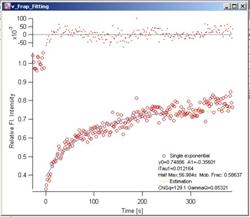
Fig. 3-9
Phair – Single Exponential Red
dots are the FRAP data. Light blue curve is the fitted FRAP curve. Decay data
are not shown. Yellow dotted curve shows the estimation curve (see 2.x for
details). At the top of the graph, fitting residuals are plotted.
3.3.8 Fitted parameters will be
indicated in three different places. (1) Within the graph as legends (2) In the
history window (3) In the fit Panel. The results printed in the history window
looks like figure 3-10.
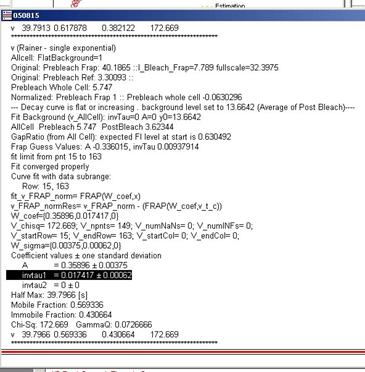
figure 3-10
GammaQ must be first checked to
evaluate the fitting (see 2.7). If gammaQ value is
less than 0.01, than you must redo fitting either by
(1)
Limit the data range for fitting by checking ‘Limit’ check box within the Fit Panel (see fig3-6) and input time
points for a desired range. After setting start and end time points, press
‘Fit!’ button again. Alternatively, you can also weight a certain range if you
check ‘Weighting’. This will
consider the specified range to be 10 times more important than the other
parts, in terms of Chi-squared value.
(2)
Use other fit models and compare gammaQ value. Larger value is better.
If you have a sufficiently large
gammaQ, then results can be used as your data. The last line in the history
window is tab delimited in the order of
experiment name – half max – Mobile
fraction – immobile fraction – Chi-Square.
You can copy and paste the line to
spreadsheet software such as Excel, and each parameter will be pasted
separately to single cells.
3.4.1. To print out graph and fitted
parameters, do Frap calc à Prepare Layout. This will
create a window called “layout” containing both the graph and the results for
printing out.
3.4.2. To export fitted parameters as
formatted text file, do Frap calc à Export Results as a text file.
3.4.3. To export frap data as
formatted text file, do Frap calc à Export Data as a text file.
3.4.4. To do both 3.4.2. and 3.4.3,
do Frap calc à Export Results and Data as a text file.
3.5 Batch Analysis (added: Feb. 2010)
When you have many FRAP experiments done
with same condition, you could average those curves and get a better idea of
more general recovery curve. In addition this has an advantage: standard
deviation at each time point could be calculated and these deviation values
could be used as parameter for curve fitting. Less standard deviation would
mean that those points are more reliable then the points with larger standard
deviation. During fitting, such difference in reliability of the measured value
could be reflected to the process of curve optimization (see chi-squared equation, 2-30) Points with less
standard deviation are typically at the initial phase of recovery, and these
points will be considered more important during the fitting.
IMPORTANT: for
averaging curves, you need to do experiments with same timing. Otherwise, batch
Fitting (see below) works and you get a list of parameters, but the program
stops when it detects that acquired time points are various in the data set.
You could use calculated parameters for each curves, maybe average them for summarizing your
results.
To do batch analysis, each FRAP data
must be saved in separate files in a same folder (currently files should be
tab-delimited text file, and the data order should be single channel Zeiss
Time-FRAP-AllCell-Base: for additional formats, please ask Kota). Then do the
following:
1. FRAP calc -> batchprocessing --beta-- -> Batch import... In
the window that pops up, choose a folder where text files with FRAP data are
stored and click OK. Then data will be loaded automatically. For each curves, a
table with data and a corresponding plot appears. During the import, a list of imported
experiment names (names generated from the file names) is created. This list is
hidden, but you could see the list by typing the following line in the command
window.
print Gfilelist
2. Then FRAP calc -> Fit Panel... In the panel, set the bleaching time
point. This bleaching time point will be commonly used for all curves. You
could already fit individual curves, but if you are in a hurry, you could
proceed to step 3.
3. Finally, FRAP calc -> batch processing --beta-- -> Batch Fit. You will
see that each individual data is curve-fitted first. Then a plot with averaged
curve appears at last. Error bars are added to the curve, so it should be
visually distinguishable from other plots. Fit results (parameters) are shown
in the plot, history and at the bottom of panel.
4. In the panel, you could change the
fit model and/or normalization methods, just like singe curve fitting in the
panel. Then you could do the "batch fit" again to test the goodness
of fit of different models.
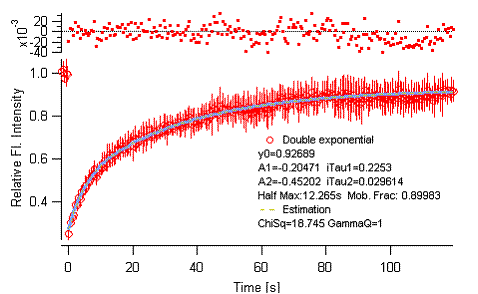
Figure 3-11.
Batch analysis, averaged curve fitting.
A. Data Format Codes
+
1111 () 0 FRAP + Whole Cell + Reference
+ Base
+
1101 () 1 FRAP + Whole Cell + Base
-
1011 () 2 FRAP + Reference + Base
-
1100 () 3 FRAP + Whole Cell
+
1010 () 4 FRAP + Reference
- 1001
() 5 FRAP + Base
+
1000 () 6 FRAP
B. Fit Method Codes
0:
Rainer - single exponential
1:
BackGroundMultiply-single exponential
2:
BackGroundMultiply-double exponential
3:
4:
(back multiply Ellenberg diffusion)
5:
(back multiply Soumpasis)
6:
Double Normalization, single exponential
7:
Double Normalization, double exponential
8:
Double Normalization Ellenberg
9:
Double Normalization Soumpasis
C. Point Number and its
content in FittingParameterWave
0: fit
method
1: normalized
2: width
3: background
exists
4: flatback
5: BackGround_Timpoint0
6: GapRatio
7: I_prebleachBack
8: I_prebleachFrap
9: I_bleachfrap
10: backamplitude
11: backtau
12: backy0
15: WholeCell_Exists
16: Ref_Exists
17: Base_Exists
Ellenberg, J., Siggia, E. D., Moreira, J.
E., Smith, C. L., Presley, J. F., Worman, H. J. and Lippincott-Schwartz, J.
(1997). Nuclear membrane dynamics and reassembly in living cells: targeting of
an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 138, 1193-206.
Jacquez, J. A. (1972). Compartmental analysis in biology and
medicine: Elsevier.
Phair, R. D., Gorski, S. A. and Misteli, T. (2004). Measurement of
dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol 375, 393-414.
Soumpasis, D. M. (1983). Theoretical analysis of fluorescence
photobleaching recovery experiments. Biophys
J 41, 95-7.
Sprague, B. L. and McNally, J. G. (2005). FRAP analysis of binding:
proper and fitting. Trends Cell Biol 15, 84-91.
Sprague, B. L., Pego, R. L., Stavreva, D. A. and McNally, J. G.
(2004). Analysis of binding reactions by fluorescence recovery after
photobleaching. Biophys J 86, 3473-95.
Acknowledgements
I
thank Martin Offerdinger (Div. Cell Biology, Biocenter Medical Uni. Innsbruck)
for finding out mistakes in formula (2-3) for the normalization of diffusion
recovery (but I should tell you, the original code was correct, it was only the
formula in this document!). I also thank
Jim Prouty (Wavemetrics) for correcting the code for IgorPro ver6.0.