CMCI
Missions
Kota Miura
Table
of Contents
1. Introduction
2. The Key Concept
of the Imaging Centre: “Systems Topobiochemistry”
3. Activities of the
Imaging Centre
1.
Introduction:
A
combination of modern microscopy and digital image processing
techniques has
radically changed biological research. We now visualize molecular
interactions
and structural dynamics as digital images to study the biological
system.
Extraction of numerical information out of three-dimensional space
within
organisms became a novel and important feature with the technological
advancements. Computer
simulation has become a realistic approach for biologists, as we can
now
simulate molecular reactions and mobility as the direct extrapolation
of actual
digital image sequences acquired through wet lab experiments. The Centre for
Molecular and Cellular Imaging (CMCI; in the following, The Imaging Centre)
will be the
key for enforcing top-quality science at EMBL in this direction. I will
discuss
the concept for this task and then will clarify the prospective
activities.
2. The
Key
Concept:
“Systems
Topobiochemistry”:
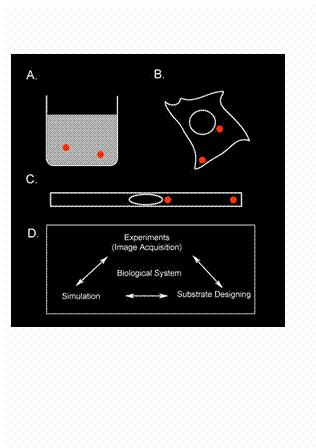
 During the 20th century, biochemistry has
accumulated an enormous amount of information on how biological system
components are functioning. Very recently, advances in fluorescence
microscopy
& imaging are pushing the knowledge forward by adding spatial
dimension as
a new parameter. I call this new area of biochemistry the “Systems
Topobiochemistry”
*1 for the following reason: In traditional biochemical
experiments
on enzyme kinetics, reactions have been studied using solutions
containing even
distributions of enzymes and substrates to characterize details of the
enzyme
function (Fig.1 A). Spatial information has been ignored in these in vitro systems partly also due to the
simple lack of technology. It allowed a clear and straightforward
analysis of
the results by decreasing the number of parameters and unknowns.
However, it has
the critical draw back that in actual biological systems, spatial
position and
local environment are key parameters contributing to
many biochemical reactions (Fig.1 B). Today, with the advancement of
microscopic and imaging technologies, the visualization of temporal
changes in
three-dimensional space has become possible. This enables the
measurement of
molecular reaction kinetics and dynamics in vivo and in situ.
Various fluorescence microscopic techniques and novel probes based on
fluorescent
proteins have evolved in last two decades. Molecular dynamics can now
be
analyzed in the natural context of the biological system.
During the 20th century, biochemistry has
accumulated an enormous amount of information on how biological system
components are functioning. Very recently, advances in fluorescence
microscopy
& imaging are pushing the knowledge forward by adding spatial
dimension as
a new parameter. I call this new area of biochemistry the “Systems
Topobiochemistry”
*1 for the following reason: In traditional biochemical
experiments
on enzyme kinetics, reactions have been studied using solutions
containing even
distributions of enzymes and substrates to characterize details of the
enzyme
function (Fig.1 A). Spatial information has been ignored in these in vitro systems partly also due to the
simple lack of technology. It allowed a clear and straightforward
analysis of
the results by decreasing the number of parameters and unknowns.
However, it has
the critical draw back that in actual biological systems, spatial
position and
local environment are key parameters contributing to
many biochemical reactions (Fig.1 B). Today, with the advancement of
microscopic and imaging technologies, the visualization of temporal
changes in
three-dimensional space has become possible. This enables the
measurement of
molecular reaction kinetics and dynamics in vivo and in situ.
Various fluorescence microscopic techniques and novel probes based on
fluorescent
proteins have evolved in last two decades. Molecular dynamics can now
be
analyzed in the natural context of the biological system.
Together
with the advancements in light microscopy technology and development of
novel
probes, two further techniques become increasingly important for the
understanding of biological systems in terms of Topobiochemistry:
computer
simulation *2 and substrate designing.
Computer simulation: Computer simulation is becoming a
powerful
technique to evaluate the image-based experimental results and its
interpretation of biological system. Following the analysis of image
sequences
from a real experiment, models of the molecular reactions and dynamics
can be
generated and the reaction or movement process can be simulated in silico. The experimental results may
suit as an initial value, from which a parameter space can be explored
and
compared with the actual experiment. The simulation results will be fed
back
into the design and refinements of further experiments.
Substrate Designing: Taken that the computer simulation is
becoming an
important part of the biological research, and that it is directly
reflecting
the actual experiments, initial and boundary conditions in the actual
experiments will be a key for the preciseness of modeling and
simulation. I
consider geometry as the most important initial and boundary condition,
which
in a biological system is typically complex and irregular. One way to
manage the
problems arising from irregular and complex initial and boundary
conditions is
to perform a geometrical transformation of the image-based experimental
results
for the simulation. A more direct and honest way to set a proper
initial- and boundary
conditions is to experimentally control the geometry of the
experimental system
through engineered environments (Fig.1 C). By means of various nano- and micro- fabrication techniques my
colleagues and I
have recently succeeded to control the cell shape and thereby also to
influence
the shape and position of intracellular structures such as the Golgi
apparatus
and mitotic spindle. Substrate designing approaches will become a
standard
protocol for imaging in a near future, because they allow highly
parallel image
acquisition of many accurately positioned and identically shaped
samples under
identical environmental conditions. Moreover, as it has been shown that
the
structure of the biological system is directly related to its function,
shape
has to be tightly controlled throughout the experiments. Substrate
design will
be an essential part of the Systems Topobiochemistry.
To summarize,
Systems Topobiochemistry consists
of three cutting-edge technologies (Fig.1 D): (1) visualization and
analysis of
molecular reactions while maintaining their spatial context within the
system, (2)
modeling and computer simulations of observed results within
multidimensional
space and (3) substrate designing to control the boundary conditions of
the
system. Top-biochemistry will be a powerful way to study the biological
systems
in this century and will be the central concept of the Imaging Centre
at EMBL.
*1 The word “Topobiochemistry” is a branch-out of the
“Topobiology”
concept proposed by Gerald Edelman as a title of his book in 1988.
Systems Topobiochemistry
is a fusion of Systems biology and topobiochemistry.
*2 See also a
recent review
article: Bork and Serrano (2005), “Towards Cellular Systems in 4D” Cell
Vol.121
p507-509.
System
topo-biochemistry at
other scales
The
essential concept of topobiochemistry is
applicable at
different scales within a biological system. Its central dogma that
spatial information
is directly related to function holds also for other
systems/environments such
as super-molecular complexes, organelles, cells and multicellular
structures. I
have been recently involved in studies of intracellular organelle
genesis and
transport dynamics as well as analysis of cell movements within tissues
in
developmental biology. At both these scales, digital image processing
enables
the analysis of spatial parameters which together with computer
simulations and
control of the structural environment has yielded high-level analytical
interpretations.
3. Activities of the Imaging Centre:
The
major task of the centre will be the development and/or refinements of
analytical techniques and tools for actual research projects. At the
same time,
spreading some of the basic knowledge to the beginners in imaging is
essential
for the success of topobiochemistry. For
this
purpose, the activity of the Imaging Centre will employ five different
types of
activity. (1) Training and Consulting on Image Processing, (2)
Collaborative
Research, (3) Software Integration and Development, (4) a Core Project
and (5) Generation of links between Light Microscopy,
Electron Microscopy and Structural Biology:
(1)
Training and Consulting on Image Processing:
As a result of the rapid increase in applications of image
based technologies, today’s biologists require some basic knowledge on
image
processing. This is not limited to the researchers who are involved in
the advanced
image acquisition and analysis, but also for those who do basic
processing of
simple single digital image files. Furthermore, “Imaging literacy” has
rapidly become
important in the interpretation and evaluation of scientific papers and
presentations,
though it is rarely taught in the course of biomedical or biophysical
studies. Despite
it has been realized some years ago by EMBL’s
scientific community that there is a strong demand for training on
image
processing, up to now support has been rather un-organized through
various
groups and people. To fill this gap, Advanced Light Microscopy Facility
(ALMF)
has been functioning as a public resource for information on microscopy
related
imaging and education. However, digital image processing and analysis
is a
broad academic field involving physics, mathematics and computer
science
knowledge and demands intensive care to be kept up to state. The
Imaging Centre
will be responsible for the task of education on digital image
processing and
at the same time build and maintain a knowledge base in this complex
area.
The aim of educational programs should be to enable
individual researchers to complete a major part of their image
processing jobs
independently after an initial consultation on the project. For
difficult
problems that require in-depth involvement of Image Centre members and
customization of the imaging software, in-house collaboration will be
considered.
(2)
Collaborative Research - Customized
analysis - :
In
most of the research projects that involve image processing and
especially
analysis as an important part of the study, customization of the
imaging
processing will be necessary. Functions native to the conventional
software in
many cases do not give satisfactory results in pioneering research
projects,
since the originality of the analytical method is directly related to
the
originality of the research outcome. For these reasons, collaborative
researches
for in-depth image processing and analyses will be an important
activity of the
Image Centre.
(3)
Software Integration and Development:
Many
researchers are currently using digital imaging microscopy to study
molecular
interactions, mobility and dynamics. Different non-commercial and
commercial imaging
software is available for these purposes, but none of them can be
always satisfactory
for individual research projects. The weak point of the
publicly-available
software is that they are too general to meet the originality of the
individual
research project. For example, tracking of organelles and movement
velocity can
be calculated with the commercial software package MetaMorph. However
the success
will be limited to those few cases when ideal shape and a high signal
quality
of the target organelles can be provided. In addition, in-depth
analysis of the
dynamics, which may provide rich information on the molecular
interaction
details, is typically hardly available. However, it may be possible to
extract this
information through both, consulting on the optimal imaging conditions,
such as
on the spatio-temporal resolutions
amenable to the
analysis algorithm used, and customization of the analysis program.
Therefore
the problem in imaging software development is the contradiction
between the
generality of public software and the originality of each research
project.
This contradiction can never be solved completely. However there are
ways to
circumvent and ease the contradiction for efficient support of
innovative
scientific projects. The solution could be provided in two ways:
software survey
and software developments.
Software
survey: Non-commercial and
commercial imaging software will be evaluated for available
functionality.
These functions will be compared for their ability and missing
functions in
different software packages will be listed. An outcome of this survey
will be
recommendation lists of software for each specific research purpose.
Another
outcome will be information on types of software, including those
already developed
by EMBL staff and used at EMBL. To ease the communication between users
of
common software, this information will be presented on the web.
Software Development: The key concept for software development
will again
consist in the “Topobiochemistry”, the concept proposed above; though
the term
is new, this concept has been practiced by biological researchers
before. As an
example, the following set of image analysis techniques is regularly
used by
molecular, cell and developmental biologists at EMBL at different
object scales.
Technique
scale
Single Particle
Tracking:
nm
FRAP, Optical
Flow
Estimation
µm
Single Organelle
/ Cell
tracking µm to
mm
All
of these algorithms are currently available in different software
modules
programmed in different ways, some of them are custom made, and others
are publicly
available. What is missing is the integration of knowledge and
techniques at
EMBL. Even for the same technique, we see different groups using
different
software, mostly because of historical but not scientific reasons.
Moreover, although
the scale is different, the techniques listed above share their aim in
analyzing the spatial information through digital imaging. All these
different
techniques can be integrated in the concept of “Topobiochemistry”:
measurement
of spatial dynamics under defined initial and boundary conditions and
computer
simulation directly related to the measurements. Accordingly, software
will be
developed as a part of a topobiochemistry
package.
Having been involved in a variety of
research
projects during the recent years, I realized that in most of the cases
the
native functions of imaging software were modified or customized
scripts and
macros had to be created to achieve completely satisfying analysis
results.
From this experience I propose that one of the major goals during the
initial
three years of the Imaging Centre shall be to transform algorithms and
programs
available at EMBL into a general software package. In this way, a
general
toolbox for biological researchers in EMBL should be developed; similar
attempt
already exist in other science institutions, for example the software
package
MIPAV (NIH, USA) mainly designed to supply the
researchers’ needs at NIH and their hard-ware instrumentations
(Microscopes).
Development
of novel imaging plug-ins and software will be done mainly in two
steps. Primordial
algorithms will mainly be designed by scientific members in close
collaboration
with the researchers. At this stage advice form software engineers will
be helpful
to identify known algorithms and increase the efficiency of calculation
processes. Afterwards these primordial pilot programs will be handed to
the software
engineers to be transformed into user-friendly software or plug-ins for
a wider
public use. Two step development procedures are required since
innovative ideas
appear at the interface of biology experiments and image processing.
These
innovative ideas must be first tested and confined through actual
research.
Problems in the algorithms and required details for the program become
also
clear during this first phase. Serious developments by programmers will
then be
the second step, with intensive discussion and information exchange
between
scientific members and engineering members.
(4)
Core projects:
An
independent core project will be carried out in the Imaging Centre. The
core
project will aim at the exploitation of pioneering approach in the
digital
image processing in biological research, to extend the topobiochemistry
by the development of cutting-edge methodology, algorithms and
technology. In
addition to the research outcome, an independent core research project
will
motivate, stimulate and maintain the pioneering atmosphere of the team.
In
addition, the core project could be helpful in developing the future
carrier of
the members becoming top scientists and engineers.
(5)
Imaging Links between Light Microscopy, Electron
Microscopy and Structural Biology:
In
deep relation to the software integration and developments, knowledge
links
between light microscopy, electron microscopy and structural biology
will be a
valuable resource for advancing the imaging technology in EMBL. In all
these
areas digital imaging has become essential for displaying the results.
Pioneering trials are being made not only to display, but also to use
more
advanced imaging methods to quantify the localization of molecular
complexes in
a resolution higher than that of the cell biological approach. To share
such information,
efforts to exchange imaging knowledge in different fields will be made
by close
communication with these fields, such as by regular seminars centered
on
imaging technologies and by inviting internal/external speakers. Inside
EMBL,
the Image Centre will be in a close contact with other Imaging related
groups
in each programme and especially with the
ALMF to facilitate
the exchange of the hardware information and image processing
knowledge.

 During the 20th century, biochemistry has
accumulated an enormous amount of information on how biological system
components are functioning. Very recently, advances in fluorescence
microscopy
& imaging are pushing the knowledge forward by adding spatial
dimension as
a new parameter. I call this new area of biochemistry the “Systems
Topobiochemistry”
*1 for the following reason: In traditional biochemical
experiments
on enzyme kinetics, reactions have been studied using solutions
containing even
distributions of enzymes and substrates to characterize details of the
enzyme
function (Fig.1 A). Spatial information has been ignored in these in vitro systems partly also due to the
simple lack of technology. It allowed a clear and straightforward
analysis of
the results by decreasing the number of parameters and unknowns.
However, it has
the critical draw back that in actual biological systems, spatial
position and
local environment are key parameters contributing to
many biochemical reactions (Fig.1 B). Today, with the advancement of
microscopic and imaging technologies, the visualization of temporal
changes in
three-dimensional space has become possible. This enables the
measurement of
molecular reaction kinetics and dynamics in vivo and in situ.
Various fluorescence microscopic techniques and novel probes based on
fluorescent
proteins have evolved in last two decades. Molecular dynamics can now
be
analyzed in the natural context of the biological system.
During the 20th century, biochemistry has
accumulated an enormous amount of information on how biological system
components are functioning. Very recently, advances in fluorescence
microscopy
& imaging are pushing the knowledge forward by adding spatial
dimension as
a new parameter. I call this new area of biochemistry the “Systems
Topobiochemistry”
*1 for the following reason: In traditional biochemical
experiments
on enzyme kinetics, reactions have been studied using solutions
containing even
distributions of enzymes and substrates to characterize details of the
enzyme
function (Fig.1 A). Spatial information has been ignored in these in vitro systems partly also due to the
simple lack of technology. It allowed a clear and straightforward
analysis of
the results by decreasing the number of parameters and unknowns.
However, it has
the critical draw back that in actual biological systems, spatial
position and
local environment are key parameters contributing to
many biochemical reactions (Fig.1 B). Today, with the advancement of
microscopic and imaging technologies, the visualization of temporal
changes in
three-dimensional space has become possible. This enables the
measurement of
molecular reaction kinetics and dynamics in vivo and in situ.
Various fluorescence microscopic techniques and novel probes based on
fluorescent
proteins have evolved in last two decades. Molecular dynamics can now
be
analyzed in the natural context of the biological system.